Preview
Creation Date
4-19-2016
Description
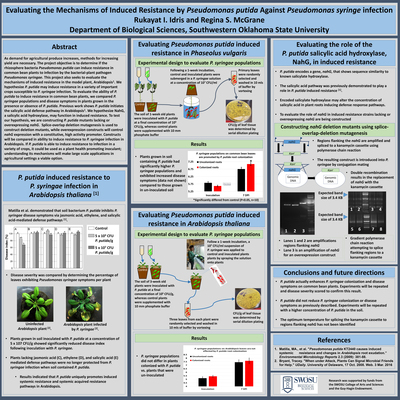
As demand for agricultural produce increases, methods for increasing yield are necessary. The project objective is to determine if the rhizosphere bacteria Pseudomonas putida can induce resistance in common bean plants to infection by the bacterial-plant pathogen Pseudomonas syringae. This project also seeks to evaluate the mechanisms of induced resistance in the model plant, Arabidopsis1. We hypothesize P. putida may induce resistance in a variety of important crops susceptible to P. syringae infection. To evaluate the ability of P. putida to induce resistance in common bean plants, we compared P. syringae populations and disease symptoms in plants grown in the presence or absence of P. putida. Previous work shows P. putida initiates the salicylic acid defense pathway in Arabidopsis1. We hypothesize NahG, a salicylic acid hydroxylase, may function in induced resistance. To test our hypothesis, we are constructing P. putida mutants lacking or overexpressing nahG. Splice-overlap deletion mutagenesis is used to construct deletion mutants, while over-expression constructs will control nahG expression with a constitutive, high activity promoter. Constructs will be compared in ability to induce resistance to P. syringae infection in Arabidopsis. If P. putida is able to induce resistance to infection in a variety of crops, it could be used as a plant health promoting inoculant; understanding its mechanisms will make large scale applications in agricultural settings a viable option.
